

1 experimentally studied protein
0 sequences in Swiss-Prot
2,941 unique sequences in UniRef100
Bacterial and some fungal l‑asparaginases
Homo dimeric structure
Containes the Rhizobium etli "type V" l‑asparaginase isoenzyme
Clan 3: Family 3
| Fam ? Class - Clan - Family | Alt ? Alternative historical name / classification | AN ? UniProt accession number | Name ? UniProt entry name, only given here for Swiss-Prot entries | EC | Organism | Cell-Loc | AAs | Structure | PDB | Km i for Asn [mM] | Vmax i for Asn [μmol/min/mg] | Kcat i for Asn [s-1] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-3-3 | ReAV | Q2K0Z2 | 3.5.1.1 | Rhizobium etli | - | 367 | Homo dimer | 7OS6 PDBs | 4.2 | - | 438 |
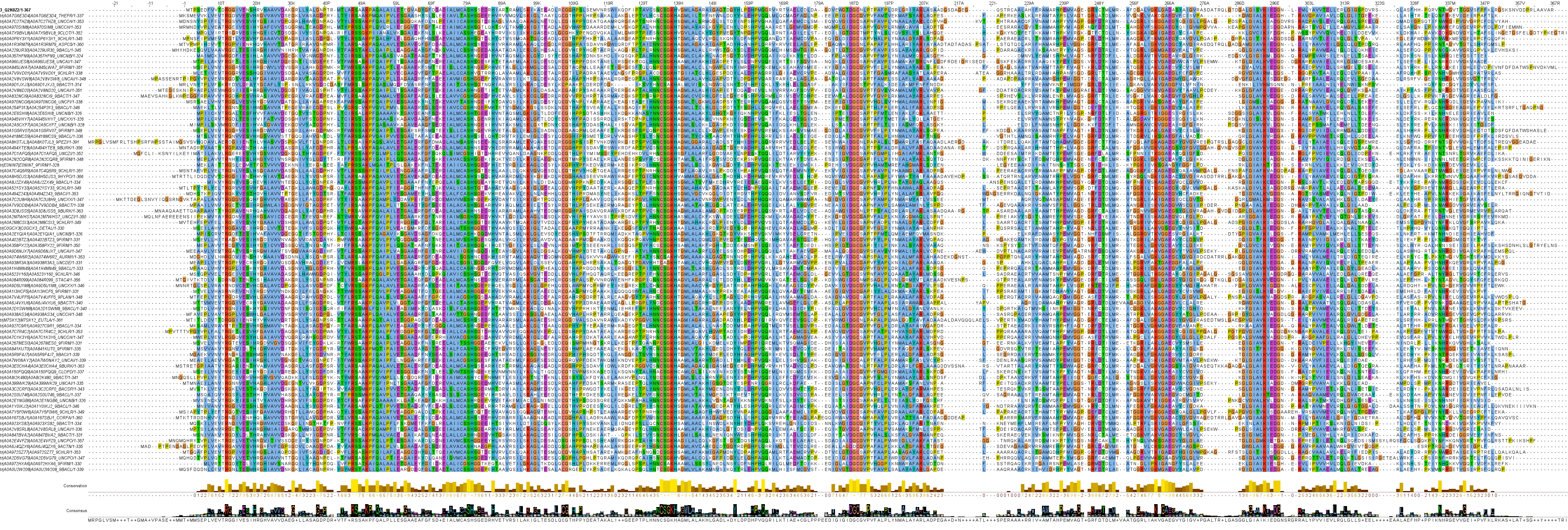
Clan 3 and Family 3 have l‑asparaginases of both bacterial and fungal origin. The crystal structure of the inducible thermolabile allosteric Rhizobium etli ReAV (Q2K0Z2) has been solved, a homo dimer with a millimolar Km for l‑asparagine.
1Varadi, M et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Research (2024). Licensed under CC BY 4.0.
2Suzek, B.E. et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics (2007). Licensed under CC BY 4.0. Added classification code to sequence headers.
