

2 experimentally studied proteins
0 sequences in Swiss-Prot
7,794 unique sequences in UniRef100
Almost exclusively sequences from bacteria in the Actinomycetota phylum
High affinity to l‑asparagine
Putative extracellular enzymes
Clan 2: Family 2

| Fam ? Class - Clan - Family | Alt ? Alternative historical name / classification | AN ? UniProt accession number | Name ? UniProt entry name, only given here for Swiss-Prot entries | EC | Organism | Cell-Loc | AAs | Structure | PDB | Km i for Asn [mM] | Vmax i for Asn [μmol/min/mg] | Kcat i for Asn [s-1] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-2-2 | - | W0KM71 | 3.5.1.1 | Nocardiopsis alba | Extracellular | 320 | - | - | 0.127 | 5.5 | - | ||
| 3-2-2 | - | Q54237 | 3.5.1.1 | Streptomyces griseus | Extracellular (putative) | 328 | - | - | - | - | - |
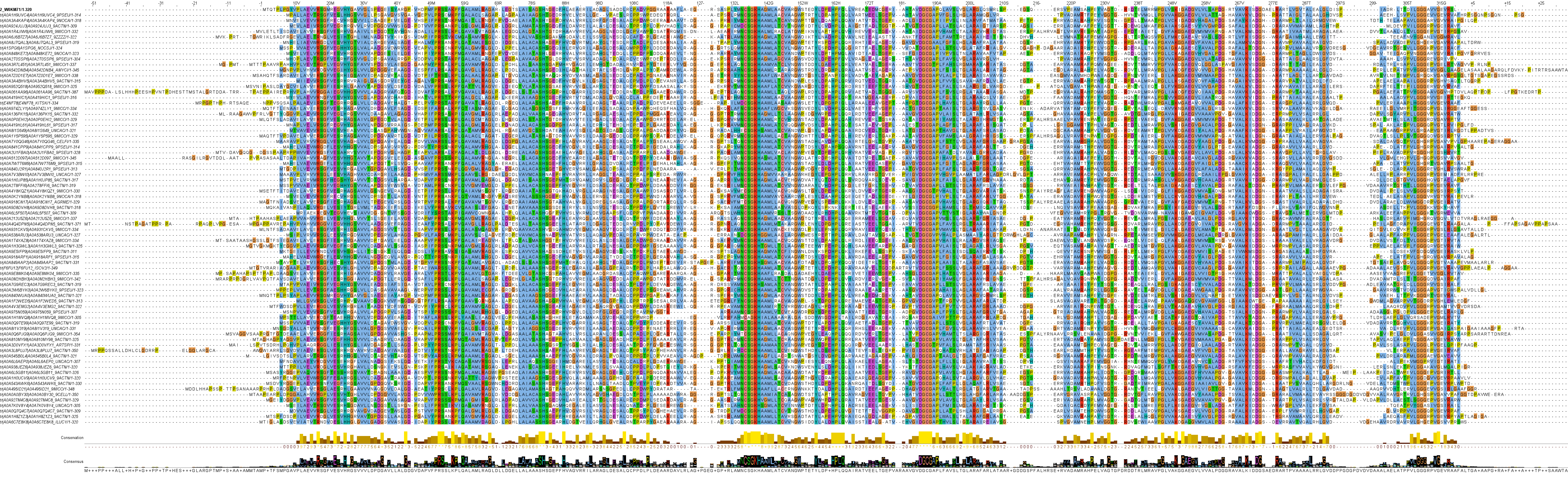
Clan 2 and Family 2 consist of sequences from the Actinomycetota phylum, like the extracellular Streptomyces griseus l‑asparaginase (Q54237) or the Nocardiopsis alba l‑asparaginase (W0KM71) with an intriguing Km for l‑asparagine in the micromolar range (not far from the Km of ErAII) and no detected l‑glutaminase activity.
1Varadi, M et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Research (2024). Licensed under CC BY 4.0.
2Suzek, B.E. et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics (2007). Licensed under CC BY 4.0. Added classification code to sequence headers.
