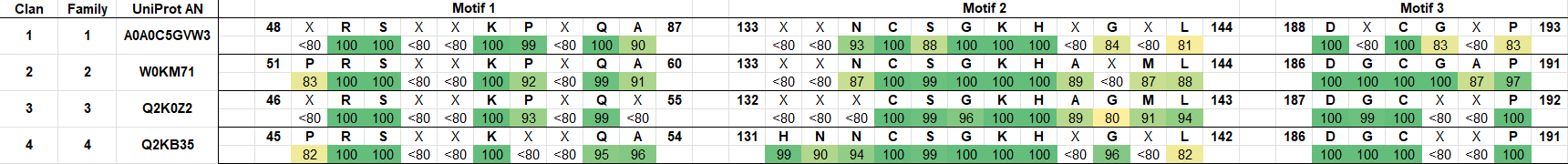
Class 3 l‑asparaginases, previously known as Rhizobium etli-type l‑asparaginases, are the smallest and most recently discovered class. Class 3 l‑asparaginases can be found in a range of bacteria, not only in the nitrogen-fixing Rhizobium etli and related bacteria, and some fungi probably due to horizontal gene transfer. It is worth noting a few archaea carrying the gene for a Class 3 l‑asparaginase were identified with BLAST.
Similarly to Class 1 and Class 2, Class 3 l‑asparaginases can show a low sequence identity to each other but the tertiary structure is well conserved in the class. Class 3 l‑asparaginases could hold untapped potential in clinical applications, as some sequences have been shown to have high affinity to l‑asparagine and undetectable l‑glutaminase activity. No Class 3 sequence with other activities could be identified in the literature. Each Class 3 clan only holds one family at the moment.