

5 experimentally studied proteins
9 sequences in Swiss-Prot
1,267 unique sequences in UniRef100
Mostly fungal l‑asparaginases
Saccharomyces cerevisiae high affinity isoenzymes

| Fam ? Class - Clan - Family | Alt ? Alternative historical name / classification | AN ? UniProt accession number | Name ? UniProt entry name, only given here for Swiss-Prot entries | EC | Organism | Cell-Loc | AAs | Structure | PDB | Km i for Asn [mM] | Vmax i for Asn [μmol/min/mg] | Kcat i for Asn [s-1] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-1-1 | - | i Sequence available in publication. No similar sequences could be found in databases. As presented, lacks the critical conserved Motif 1. | 3.5.1.1 | Cladosporium sp. | - | 370 | - | - | 100 | - | - | ||
| 1-1-1 | - | A0A4D6C6W0 | 3.5.1.1 | Cobetia amphilecti | - | 316 | - | - | 2.05 | 11641 | - | ||
| 1-1-1 | - | C5E3N2 | 3.5.1.1 | Lachancea thermotolerans | - | 375 | - | - | 11.67 | - | 312.45 | ||
| 1-1-1 | ScAII i Identical sequences: P0CZ17, P0CX77, P0CX78, P0CX79 | P0CZ17 | ASP21_YEAST | 3.5.1.1 | Saccharomyces cerevisiae | Secreted | 362 | - | - | 0.27 | 42 | - | |
| 1-1-1 | ScAI | P38986 | ASPG1_YEAST | 3.5.1.1 | Saccharomyces cerevisiae | Cytoplasm | 381 | - | - | 0.075 i K0.5, allosteric enzyme | - | - |
| Fam ? Class - Clan - Family | Alt ? Alternative historical name / classification | AN ? UniProt accession number | Name ? UniProt entry name, only given here for Swiss-Prot entries | EC | Organism | Cell-Loc | AAs | Structure | PDB | Km i for Asn [mM] | Vmax i for Asn [μmol/min/mg] | Kcat i for Asn [s-1] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-1-1 | ScAII i Identical sequences: P0CZ17, P0CX77, P0CX78, P0CX79 | P0CZ17 | ASP21_YEAST | 3.5.1.1 | Saccharomyces cerevisiae | Secreted | 362 | - | - | 0.27 | 42 | - | |
| 1-1-1 | ScAII i Identical sequences: P0CZ17, P0CX77, P0CX78, P0CX79 | P0CX77 | ASP22_YEAST | 3.5.1.1 | Saccharomyces cerevisiae | Secreted | 362 | - | - | 0.27 | 42 | - | |
| 1-1-1 | ScAII i Identical sequences: P0CZ17, P0CX77, P0CX78, P0CX79 | P0CX78 | ASP23_YEAST | 3.5.1.1 | Saccharomyces cerevisiae | Secreted | 362 | - | - | 0.27 | 42 | - | |
| 1-1-1 | ScAII i Identical sequences: P0CZ17, P0CX77, P0CX78, P0CX79 | P0CX79 | ASP24_YEAST | 3.5.1.1 | Saccharomyces cerevisiae | Secreted | 362 | - | - | 0.27 | 42 | - | |
| 1-1-1 | - | P87015 | ASPG1_SCHPO | 3.5.1.1 | Schizosaccharomyces pombe | Secreted, cell wall | 360 | - | - | - | - | - | |
| 1-1-1 | ScAI | P38986 | ASPG1_YEAST | 3.5.1.1 | Saccharomyces cerevisiae | Cytoplasm | 381 | - | - | 0.075 i K0.5, allosteric enzyme | - | - | |
| 1-1-1 | - | Q9UTS7 | ASPG2_SCHPO | 3.5.1.1 | Schizosaccharomyces pombe | Secreted, cell wall | 356 | - | - | - | - | - | |
| 1-1-1 | - | Q8NKC0 | ASPG3_SCHPO | 3.5.1.1 | Schizosaccharomyces pombe | Secreted, cell wall | 360 | - | - | - | - | - | |
| 1-1-1 | - | Q8TFF8 | ASPG4_SCHPO | 3.5.1.1 | Schizosaccharomyces pombe | Secreted, cell wall | 356 | - | - | - | - | - |
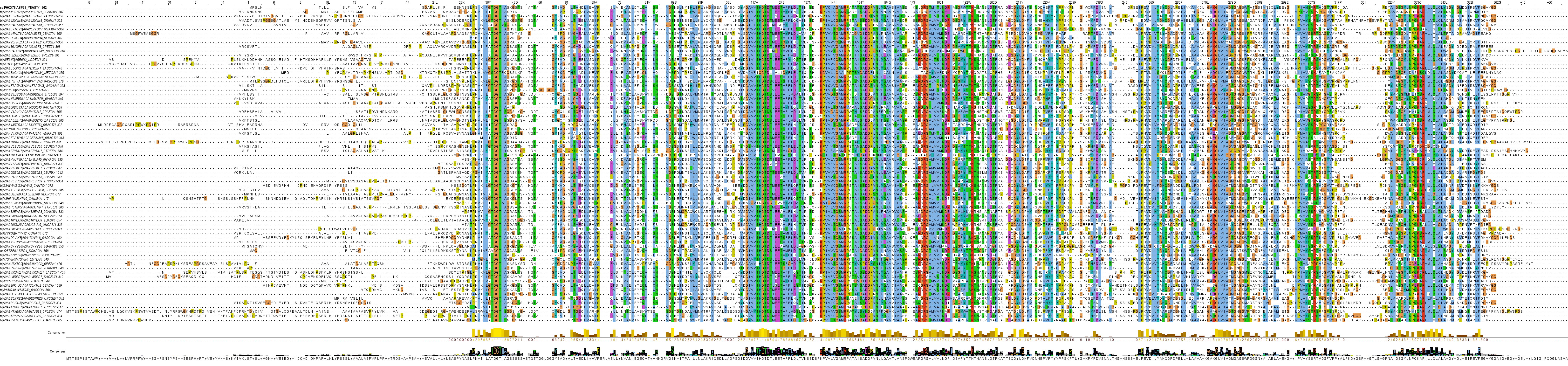
Clan 1 holds 4 families and is somewhat analogous to the historical "type II" l‑asparaginases, as the families of this clan contain "type II" sequences, including the two in therapeutical use. It also has many cytoplasmic sequences and some of these were previously considered to be "type I" l‑asparaginases.
Family 1 contains mostly fungal l‑asparaginases, including the cytoplasmic Saccharomyces cerevisiae ScAI (P38986) and the allosteric cell wall Saccharomyces cerevisiae ScAII (P0CZ17) (several copies present in genome). Both enzymes show high affinity to l‑asparagine with a Km in the micromolar range.
1Varadi, M et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Research (2024). Licensed under CC BY 4.0.
2Suzek, B.E. et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics (2007). Licensed under CC BY 4.0. Added classification code to sequence headers.